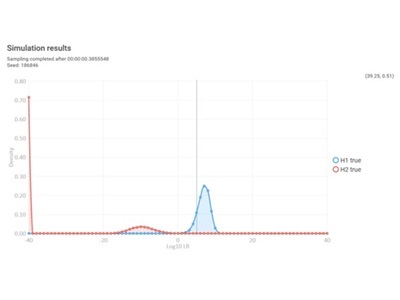
Simulated LR distributions for comparison against the trace component (contributor 3). These results indicate that the trace component of the mixture has probative value, despite having a low estimated template.
by Adam McCarthy, Associate, STRMix, United Kingdom and European Associate
Forensic DNA profiling laboratories find themselves under increasing pressure to efficiently utilize resources and apply the most modern approaches to deliver robust evaluative and intelligence information. To produce those results, a growing number of labs are accessing a combination of software solutions that enable them to complete the full workflow, from DNA analysis to interpretation and database matching.
A hypothetical homicide case demonstrates how an integrated suite of software applications can be used in a typical laboratory workflow. The case involves the recovery of a firearm used in a homicide. No person(s) of interest (POI) have been identified in the early phase of the investigation. Swabs used to sample areas of the firearm and recover biological material have been extracted. DNA has been recovered and amplified to identify any person(s) that can be linked to the incident. Capillary electrophoresis (CE) DNA profiling technology has been applied and the resulting raw data is now available for analysis.
Just to achieve these raw results, the resource, expertise, and quality system demands are significant, so any tangible investigative or evaluative outcome will benefit all interested parties. To generate quality raw data and convert it into useful intelligence quickly and efficiently, the forensic lab begins by analyzing raw CE data in the case using FaSTR DNA, an analysis software which can accept input files in different formats (.fsa/.hid/.promega) generated by different CE instruments.
FaSTR DNA uses dynamic baselining of the EPG data, detection and assignment of size-standard peaks for the accurate sizing of peaks in the allelic ladder and test sample, and size alignment of peaks detected in the allelic ladder to assist and streamline the workflow. It can also apply a set of analysis rules to distinguish artifactual from allelic peaks and carry out a Number of Contributor (NoC) estimation using a published decision tree method1. In the Review Module, a second analyst can confirm the results.
At this point, the forensic lab is able to carry out sample-to-sample or sample-to-database comparison checks, as well as full concordance testing on positive and negative controls. At analysis completion, the results can then be exported as configurable outputs.
In our hypothetical case, the recovered material has generated a DNA profile originating from a suggested three contributors. The NoC assignment in the software provides some fast, empirically derived support for this and initial visual assessment of the contributor DNA mixture proportions provide some insight into varying amounts of DNA from those contributors.
With this information in hand, the sample would benefit from analysis (or deconvolution) by probabilistic genotyping (PG) software to help in linking it to a person of interest. Unlike previous methods of DNA analysis, which depended entirely on the application of fixed stochastic thresholds and other biological parameters to manually analyze DNA samples, PG software uses a fully continuous model that makes better use of the information available within a DNA profile. It proposes hundreds of thousands of different profiles and is able to assess and weigh how closely these resemble the observed DNA mixture.
Because STRmix PG software fully integrates with FaSTR DNA, the lab uses it to run our hypothetical case example, along with any other samples in our CE analysis collection, in Batch Mode. There, analysis can be configured to compare with any available standard (or reference) samples from a POI.
STRmix outputs the useful diagnostic of mixture proportions and, in this case, the sample benefits from this refined insight. The analyst, using their experience and expertise to check this diagnostic against observed results, agrees that the mixture proportions are approximately 4:2:1.
Given the absence of any standard from a POI for comparison, the weight (being the primary output of STRmix and a representation of the likelihood of the observed (EPG) results given a tested genotype combination) can be formatted for comparison against a local or national database (e.g. CODIS).
To further elevate the intelligence information in our hypothetical case and complete the full workflow, the lab turns to DBLR, an investigative application which uses efficient algorithms to rapidly calculate millions of likelihood ratios (LRs) in DNA evidence. Because DBLR fully integrates with STRmix, the lab can import STRmix deconvolutions or single-source profiles and visualize the evidential value or carry out rapid database searches. These insights can be used to help scientists and investigators decide on resource and effort priorities. Does the sample warrant further processing? Is there sufficient information in results obtained to direct the investigation? Can these results be used to link to other cases?
The visual outputs (above photo) in DBLR also communicate “at a glance” information to interested parties. These outputs utilize expected LRs for one or many components of DNA profiles for true and non-contributors using randomly generated individuals.
In our hypothetical case, component 3 of this mixture appears suitable for comparison. Based on the distributions of expected LRs, we may expect good discrimination capacity and should be able to clearly distinguish true from false donors. The Search Database function of DBLR™ can then be used to search component 3 against saved databases in seconds.
The implication of all this is clear. Utilization of the total workflow solution, which in our hypothetical case was illustrated by combining FaSTR DNA, STRmix, and DBLR, can help to ensure that the time and expertise spent in generating quality raw data is converted into crucial intelligence and investigative information with speed and efficiency.
Reference
1. Kruijver et al. Estimating the number of contributors to a DNA profile using decision trees, Forensic Science International: Genetics 50 (2021) 102407
About the author
A former reporting officer for the Home Office Forensic Science Service, Adam McCarthy has presented expert testimony in both biological and firearms casework examinations. McCarthy joined the STRmix team in 2015 and provides representation for the group in the United Kingdom, Europe, and RoW regions. For more information, visit http://www.strmix.com.